Working with Databases in R
As a data analyst having the skills and knowledge to manage and analyze vast datasets efficiently is crucial. R programming languages has packages to help seamlessly interact with databases.

Introduction
Hello. 😇 In this post, you will be introduced to the basics of connecting to a database and retrieving data with SQL queries using the DBI package. Next, we’re not going to look at dbplyr which helps to translate dplyr code into the SQL. Don’t worry if you are not a master of SQL, a structured query language, we will look at a shortcut to generate SQL from R. Let’s dive in.
To begin, let’s start by understanding what the two main packages - DBI and dbplyr do for us.
DBI is a low-level interface that connects to databases and executes SQL; dbplyr is a high-level interface that translates your dplyr code to SQL queries then executes them with DBI.
To install these packages as well as our trusty tidyverse package, which contains essential packages like dplyr, this is shown in the code below:
install.packages(c("DBI", "dbplyr", "tidyverse"))To use these packages, we have to load them in our RStudio IDE as follows
library(DBI)
library(dbplyr)
library(tidyverse)The main operation of a database is run by database management systems, generally known for short as DBMS. These come in three basic forms - client-server, cloud, and in-process. In this article, we will be exploring the in-process DBMS, duckdb which runs entirely on your local machine, is great for working with large datasets, and can be shared with your project.
It’s time to start working with the database. We need to install and load duckdb, a package tailored for the DBMS we are connecting to, duckdb. Same name as the DBMS, that’s interesting. 🙂
install.packages("duckdb")
library(duckdb)Create a named database
We will be creating and connecting to a new duckdb database, coffee_db as follows.
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = "coffee_db")The database name was an inspiration by Mo Chen’s tutorial on YouTube. You can get the code for generating this database structure, data, and the code used for this post on my Github repo.
Let’s get a view of the tables in this database
DBI::dbListTables(con)
#> [1] "employees" "locations" "shops" "suppliers"Manage Local machine memory: Shutdown database
Remember to always shutdown your database when you are not working with it as follows:
DBI::dbDisconnect(con, shutdown=TRUE)Connect to Existing Database
To connect with an existing database in your project, use the code below.
# this connection connects to an existing database, coffee_db
con <- DBI::dbConnect(duckdb::duckdb(dbdir = "coffee_db"))Explore Database table
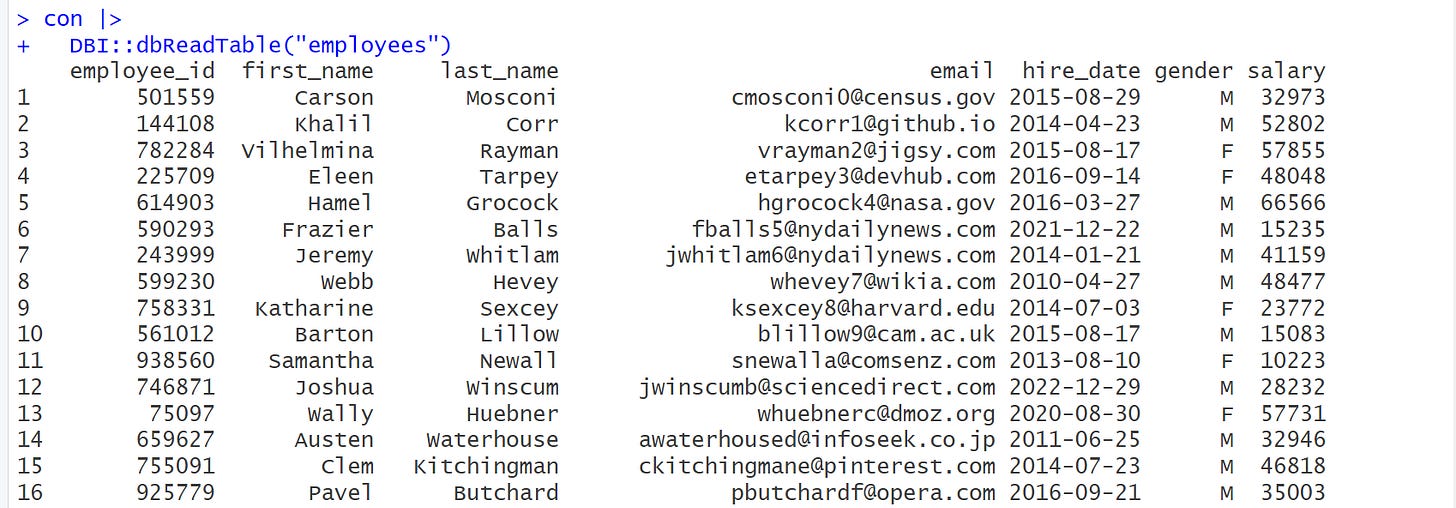
Next, we want to see the data from the employees table. 🤗 We will use dbReadTable() to retrieve the contents of the table as follows
# # view data from table.
con |>
DBI::dbReadTable("employees") Note that dbReadTable() returns a data.frame as output which doesn’t give us that nice printout. To get a more nice and informative printout, we use as_tibble() to convert it into a tibble as shown below.
# view table as a tibble in R
con |>
DBI::dbReadTable("employees") |>
as_tibble()Great job. 🫡 We have 1000 rows and 8 columns in our employees table. You can modify the code to view other tables within the coffee_db database.
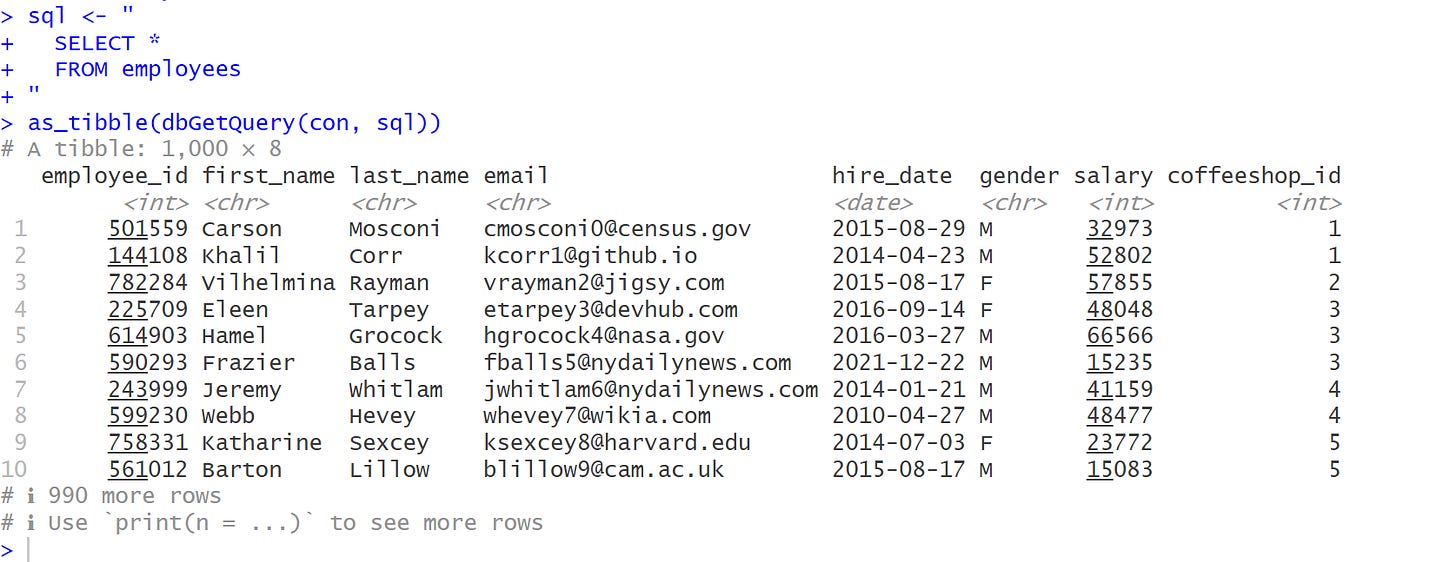
Return Table data with SQL
We can also use SQL queries to run a query on the database by calling DBI’s dbGetQuery():
# using SQL queries
sql <- "
SELECT *
FROM employees
"
as_tibble(dbGetQuery(con, sql))As you can see, this is easy for SQL enthusiasts and we do not need to convert to a tibble this way. 🤓
Great job so far.🤝 We’ve learned how to connect to a database and loaded up some data from a database table. It’s time to learn about dbplyr. Interacting with database in R basically done writing code using the dplyr package. But the database which serves as the backend does not understand code except SQL. So, the backend executes these codes definitely with the help of dbplyr which translates the codes to SQL.
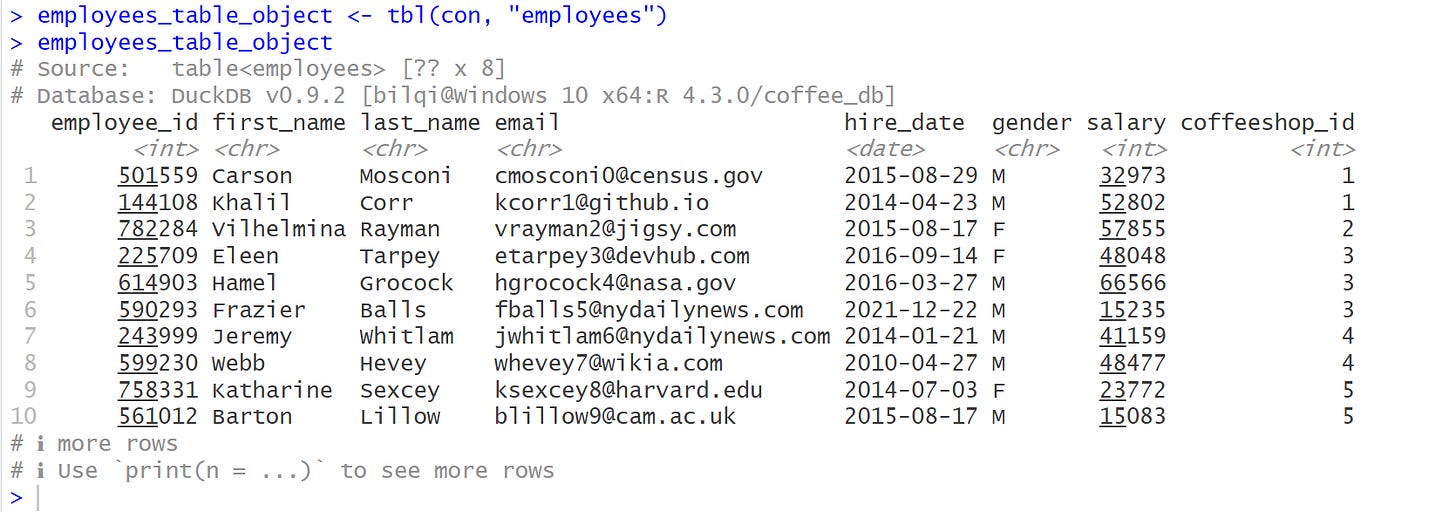
To use dbplyr to translate the dplyr code, we use tbl() to create an object that represents a database table.
# database table object
employees_table_object <- dplyr::tbl(con, "employees")
employees_table_objectFrom the output above, the structure of the object is termed lazy. That is, when you use dplyr verbs on a database object, dplyr doesn’t do any work: it just records the sequence of operations that you want to perform and only performs them when needed.
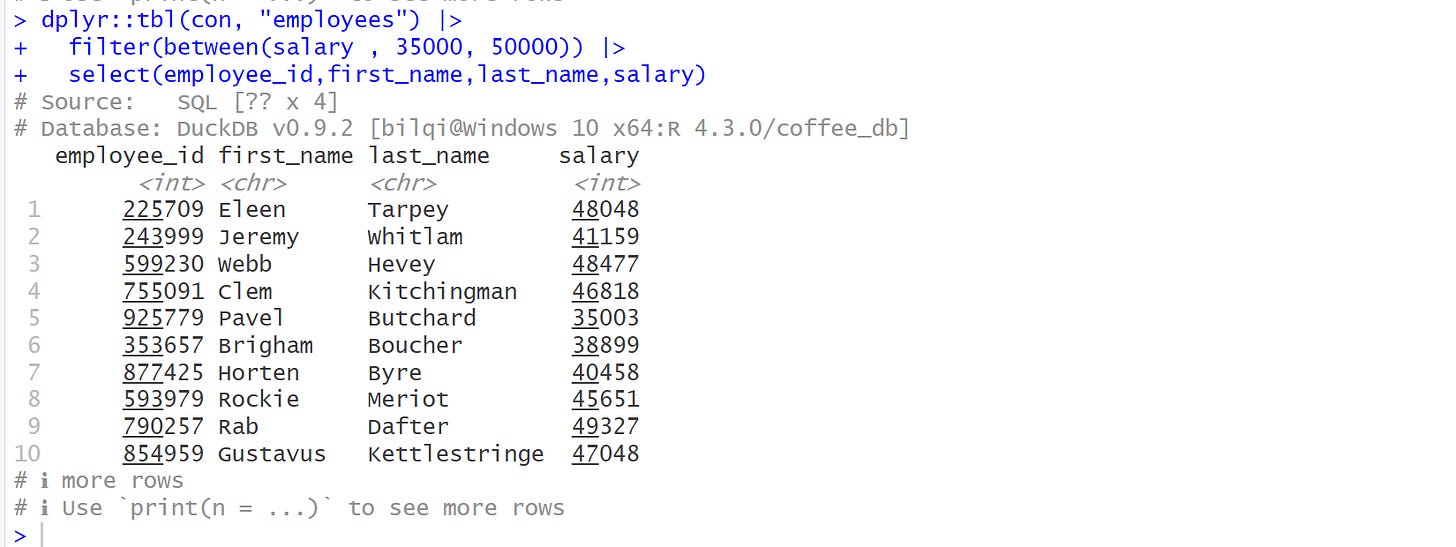
Let’s look at another interaction, using the dplyr code
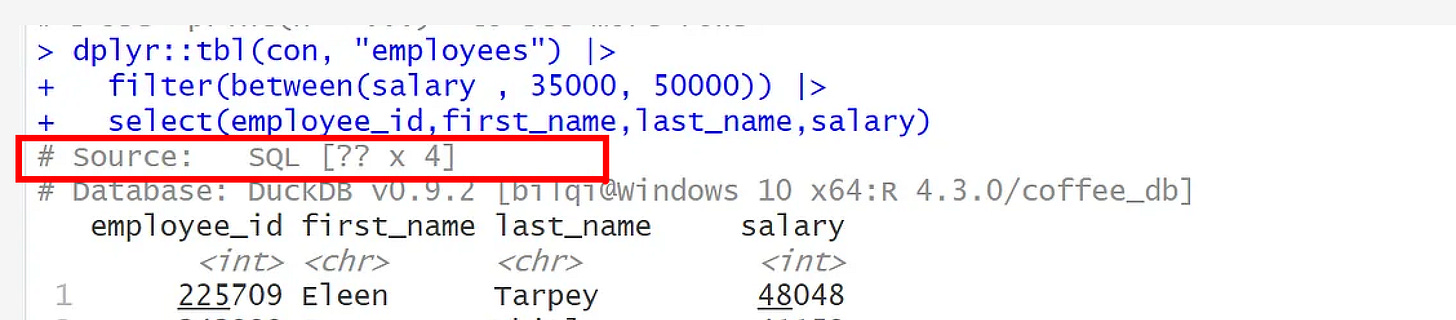
dplyr::tbl(con, "employees") |>
filter(between(salary , 35000, 50000)) |>
select(employee_id,first_name,last_name,salary)From the output, you can observe that this object represents a database query and not a table object.
You can see that it prints the DBMS name at the topshowing the number of columns as 4, while number of rows is ??, which means, it doesn’t know the number of rows. This happens because the total number of rows is computed when the complete query is executed, which typically is something we’re trying to avoid. Why? I will explain next.
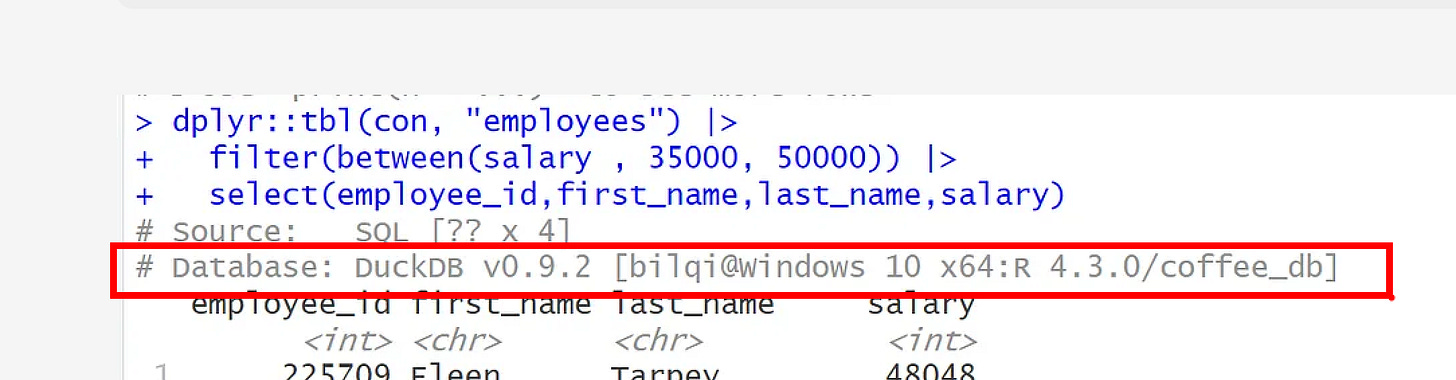
Lastly, from the printout, you can also see the database information showing the DBMS as DuckDB, version, location and name.
Now, let me answer the why question above. We can generate the SQL code by piping the dplyr function show_query() as shown below.
# Generate SQL code
dplyr::tbl(con, "employees") |>
filter(between(salary , 35000, 50000)) |>
select(employee_id,first_name,last_name,salary) |>
dplyr::show_query()Magic! 😀 No. That’s why, you do not need to sweat to learn SQL when you know dplyr! 😎 Just write some dplyr code, send it to dbplyr to translate it to SQL using the dplyr function show_query(). I have an article to show you how
Delaying the SQL query execution enables us to collect the actual data using the dplyr’s collect() function.
dplyr::tbl(con, "employees") |>
filter(between(salary , 35000, 50000)) |>
select(employee_id,first_name,last_name,salary) |>
dplyr::collect()Now, you can see that the final output is a complete tibble with 272 rows and 4 columns. Hurray! 🤩🤩🤩 We can see how taking little steps we can connect and get our required data without overburden our environment. I will summarise the steps as follows.
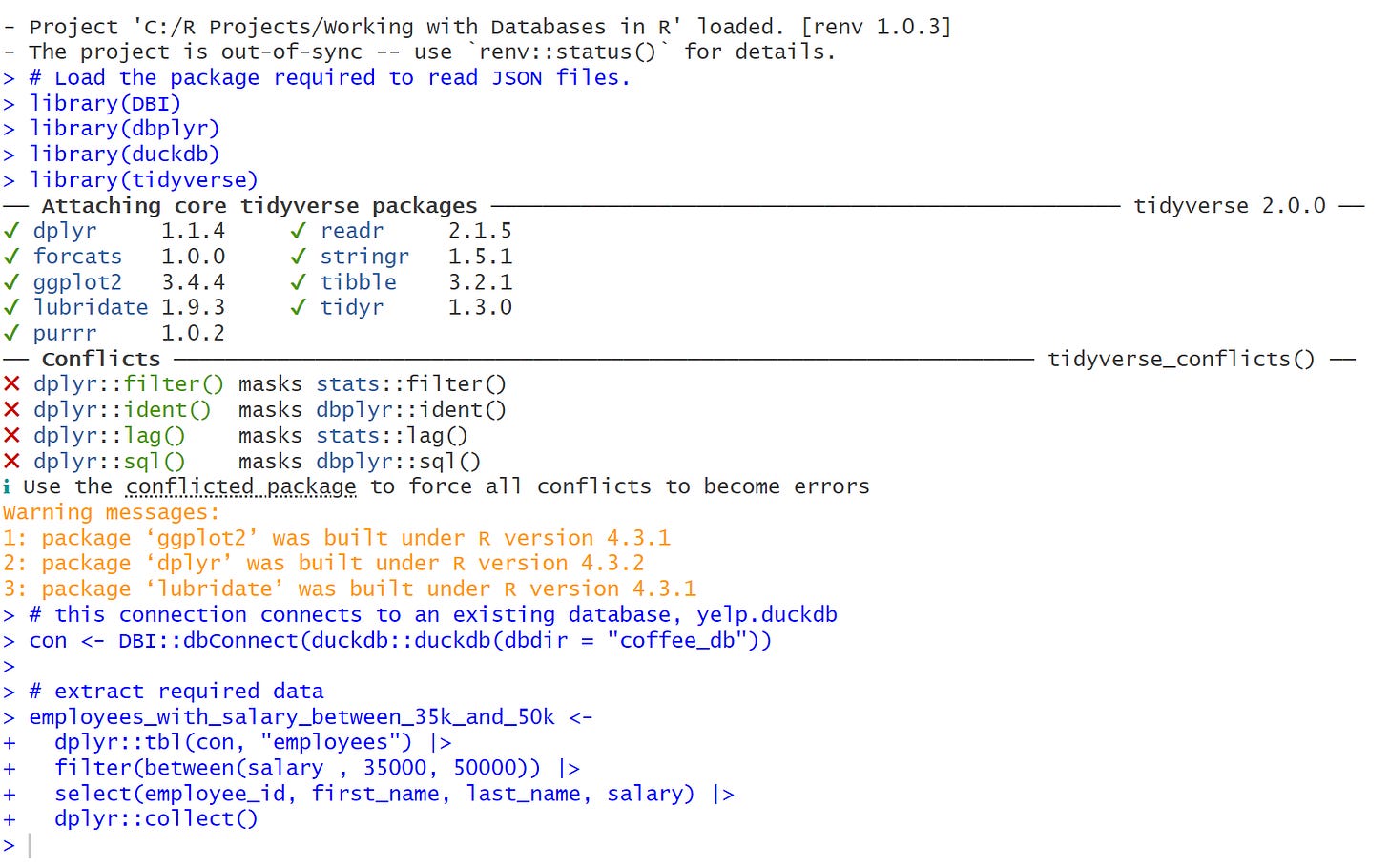
# Load the package required to read JSON files.
library(DBI)
library(dbplyr)
library(duckdb)
library(tidyverse)
# this connection connects to an existing database, yelp.duckdb
con <- DBI::dbConnect(duckdb::duckdb(dbdir = "coffee_db"))
# extract required data
employees_with_salary_between_35k_and_50k <-
dplyr::tbl(con, "employees") |>
filter(between(salary , 35000, 50000)) |>
select(employee_id, first_name, last_name, salary) |>
dplyr::collect()I want to thank you for coming with me this far. We have learned how to take just few steps to connect, interact and collect data from the database in R. I hope you enjoyed it.
Thank you.